Synthetic biologists, like Eric Klavins and his colleagues at Washington State University, attempt to engineer living things to carry out machine functions, such as computing. This group recently reported advances in making computer circuits in living cells, known as biocircuits. These circuits have the advantage of avoiding some of the messiness of living systems.
A machine is designed to accomplish a task assigned to it by humans, while evolution has built living things to survive. A machine’s parts are relatively simple, because they are built to accomplish a specific sub-task. In contrast, parts of living things can be much more complex, because the many systems in a cell have evolved alongside each other. Separate components can act in multiple pathways, and roles are filled by redundant parts with overlapping responsibilities.
Genetic Circuits
Like computers, cells process information. One way they do this is by converting the sequence of information in DNA genes into a chemically similar sequence in another molecule, called RNA. This process is called transcription. The decision to transcribe a gene can be thought of as turning a circuit on. The decision not to transcribe can be thought of as turning a circuit off. However, that analogy is an over-simplification. Unlike electrical circuits, genes are almost never completely turned off - this is known as “leakiness.” This makes them poor materials with which to build computing circuits. The Klavins group overcame this problem by using the Mxi1 protein in their experimental yeast cells. In this cell, Mxi1 is an extremely effective silencer of transcription - bringing the leakiness down to acceptable levels.
Another problem is how to connect one circuit to another. Earlier work addressed this issue using CRISPR technology. CRISPR stands for ‘Clustered Regularly Interspersed Palindromic Repeats.’ This is an immune system in some bacteria. When these bacteria survive an encounter with a virus, they keep a copy of a fragment of the viral gene sequence as a short fragment between palindromic (back to front) repeats in their DNA. When it collects a bunch of these sequences it has what is known as a cluster.
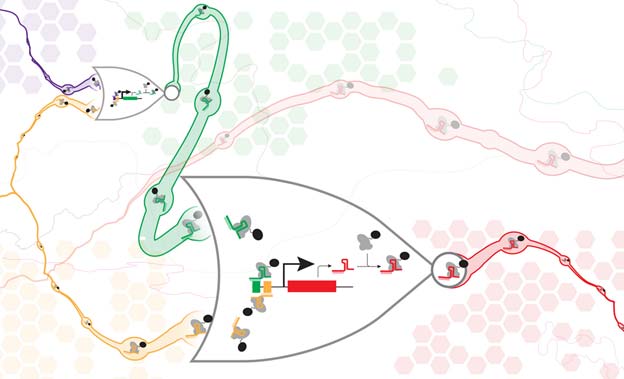
An artist’s impression of connected CRISPR-dCas9 NOR gates. Justin Vrana, University of Washington
These saved fragments are transcribed as part of ‘guide’ RNAs that direct an enzyme, Cas9, to attack these sequences if they ever see them again. Scientists can replace the guide RNA with whatever sequence they want, and then transplant the system into their favorite cell. In artificial genetic circuits you can turn off genes by blocking their transcriptional start site with a Cas9 led by a guide RNA against the circuit’s specific DNA sequence. Unfortunately, this kind of connection is leaky.
The Evans group improved on this strategy by engineering a dCas9-Mxi1 fusion protein. This protein combines the silencing strength of Mxi1 and the specificity of dCas9.
A Logical Circuit
The Klavins group engineered a ‘NOR’ gate. NOR is an operator from Boolean Logic. (Traditionally, Boolean operators are written in all caps.) Boolean logic is sometimes used in library search engines. ‘AND’ is an example of another Boolean operator. If you search in a Boolean library system and write: ‘puppies AND cute,’ you will get all documents that include both terms and you won’t get documents that refer to puppies that aren’t cute. You also won’t get documents that only refer to cute kittens (or any other cute thing, just puppies). The documents must contain both terms.
‘NOR’ could be read as ‘neither.’ The logical statement is satisfied only if both terms are negative. In a library it would give you all the documents that don’t mention ‘puppies’ or ‘cute.’ (How many could there be?)
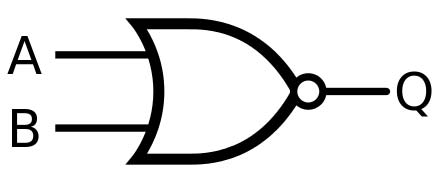
A diagram of a NOR gate (Public Domain)
A ‘gate’ is a device that operates as a Boolean operator. An electrical NOR gate has two inputs (A and B) and one output (Q). If both inputs are off, the output is on. Perhaps one input reports on the presence of ‘puppies.’ The second input could report on the presence of ‘cute.’ If both inputs report negative, the output will be on (it will transmit current). This electrical current would be a computer’s way of saying: Yes, we have no ‘cute’ or ‘puppies.’
Building It with DNA
The genetic NOR gate has a transcriptional start site for making RNA that is ‘on’ if left alone. That generates the output. However, two different dCas9 target sites are built into it. The dCas9-Mxi1 fusion protein can bind to one, or both, of these sites with two different guide RNAs. If it does, there is no output because the Mxi1 component suppresses RNA transcription. There will only be an output if both dCas9 target sites (the inputs) are empty. The RNA transcript output contains another dCas9 guide that could bind to a second NOR gate. This would connect two gates in a circuit.
More Complicated Circuits
As a matter of logic, NOR gates are one of the few gates that can be used to build all the others. For example, If the inputs are identical it becomes a ‘NOT’ gate. That is, if the two inputs are ‘cute’ into the NOR gate, it is off. This is equivalent to ‘NOT.’ The output then is ‘not cute.’
The Klavins group linked together NOR gates into more complicated circuits, as many as seven in a row. In order to measure the result, the last output is a ‘messenger’ RNA that can be translated into a protein that is easily detected. This is actually typical; the best known RNAs are messenger RNAs that are used to make proteins.
Possible Limitations
Genetic circuits won’t ever be as fast as electronic circuits. Professor Klavins writes in an email, “Electronics is much faster than the kind of biocircuits we (and many others in synthetic biology) currently make. The biocircuits we constructed harness DNA transcription in the cells to create the output of each gate upon detection of the gate’s inputs. And transcription is just a slow process…” That is to say, slow in comparison to the movement of electrons through wires or silicon.
Another problem is degradation. All information transmission has this problem. Gates of any kind cannot be strung together in an infinite series in the real world. Leakiness seems to be the main cause of signal degradation in biocircuits.
The Future of Biocircuits
According to Professor Klavins, “…the kinds of biocircuits we are making are along the lines of the logic that goes into cell differentiation, development, and morphogenesis. I want to understand how to reprogram those processes…” He is referring to the process by which cells decide what they are going to be when they grow, and how organisms build tissues and body parts. Professor Klavins has also talked about cells, modified with biocircuits, living in a tissue deciding if that tissue needs treatment and dispensing that treatment from within that tissue.
This report is a big step forward in biosynthetics, but we are still in the early stages in this field that hopes to harness living machines to produce real benefit in industry and medicine.
Top image: Example of a part of printed circuit board (CC BY-SA 3.0)
References
Langston, J., 2017. UW engineers borrow from electronics to build largest circuits to date in living eukaryotic cells. [Online]
Available at: http://www.washington.edu/news/2017/05/25/uw-scientists-borrow-from-electronics-to-build-largest-circuits-to-date-in-living-eukaryotic-cells/
[Accessed 7 June 2017].
Miles W. Gander, J. D. V. W. E. V. J. M. C. E. K., 2017. Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nature Communications, 25 May.Volume 8.
Wikipedia, 2017. NOR gate. [Online]
Available at: https://en.wikipedia.org/wiki/NOR_gate
[Accessed 7 June 2017].






No comment