How do defects that affect the upper lip and palate occur at birth?
This is a question that researchers from Yale University and the University of Connecticut set out to answer, using advanced methods and databases. The results, as reported in a May 2018 issue of the journal, Cell Reports, has indicated that the answer is a little more complicated than one involving a handful of genes controlling bodily structure formation.
The scientists found that an entirely different panel of genes and genetic interactions are more likely to be affected, in the cases of facial or palate clefts. These genes are responsible for the coordination of embryonic development as it progresses in the womb.
What is a Cleft Lip?
Clefts of the upper lip, which have also been known as “hare-lip," affect one in about 700 live births. These defects often change the shape of the lip just under the nose but may also run deeper into the palate. Some clefts can be resolved with surgery, but others are not severe enough to warrant such interventions. Researchers believe that these clefts are associated with mutations in a small number of specific genes.
However, many studies suggest that these mutations impact a relatively small number of people with the condition. Therefore, theses such as these do not explain the cause for a majority of clefts.
As cleft lips are common, it may be beneficial for those affected if the problem of how cleft lips develop, in utero, is solved once and for all.
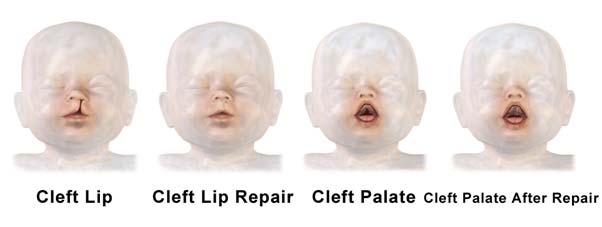
Picture showing an illustration of a child with a cleft lip and palate and its subsequent repair by plastic surgery. (Source: Wikimedia Commons)
Study of Genetics of the Cleft Lip
A team of researchers from the health research subsidiary of the University of Connecticut (UConn Health), Yale University and the Institute for Systems Genomics at Connecticut, Storrs, conducted a detailed whole-genome study to assess any other probable genetic links with the cleft lip/palate syndrome.
The team found that a cleft lip is most likely to be a disorder of the epigenetic control of embryonic expression patterns. This system is associated with tracts of the human genome not traditionally thought to be a part of ‘functional’ sequences or those that code for the actual proteins and similar molecules that make up the bones, muscles, cells and their components.
The researchers have included in their paper mechanisms such as chromatin signaling and histone modifications, which are ‘big picture’ types of genome editing, or regulation at the chromosomal level.
This epigenetic system contains “enhancers," or positive regulatory molecules that ensure that structures such as the skull build themselves in an appropriate manner. They are typically in force during the first ten weeks or so into embryonic development.
Normal, successful enhancer functions result in a normal skull and face shape. However, their impairment may lead to conditions such as cleft lips.
The Connecticut researchers assessed the histone modifications and chromatin states of human fetal tissue samples, donated at stages ranging from four to eight weeks of development, using next-generation high sequencing. They validated these findings against the 2015 Roadmap Epigenomics Consortium and 127 public genomic databases.
Genetic Assessment: The Role of Epigenetic Enhancers
The team identified thousands of chromatin state-related epigenetic regulators that were likely to influence the face and skull structure at relevant stages of fetal development.
These regulators bound a total of seven genes previously implicated in cleft lip and/or palate development. The team compared their regulators with other established models of craniofacial formation in humans and other animals. They found that many of their regulators were significantly associated with skull or face formation determination.
In addition, the scientists found numerous non-chromatin-related enhancers and repressors, which were significantly enriched, in their analysis.
This pool of potential craniofacial enhancers underwent further comparison with those identified using chromatin immunoprecipitation of epigenetic controllers, a powerful analytical technique to assess enhancers or repressors.
The researchers then compared these enhancers with data from the Consortium and the 127 databases. This led to the identification of 6,651 enhancers in total, representing just under 9% of the entirety of craniofacial structure-associated enhancers.
The enhancers had roles that included the pre- and post-translational modification of genes more directly involved in craniofacial development. The pieces of DNA were also associated with the H3K27ac locus of histone modification. Additionally, a relationship was observed with several different chromosomes including those of 1, 10, 15, 17 and 22.
The group of scientists compared their nominated enhancers with those known to be associated with genetic conditions that have resulted in cleft palates, in the past. This part of the study showed some positive matches. For example, some enhancers had been implicated in a skull-structure disorder known as Van Buchem disease. Therefore, it was concluded that the enhancers identified in this study were likely to be associated with the risk of facial clefts.
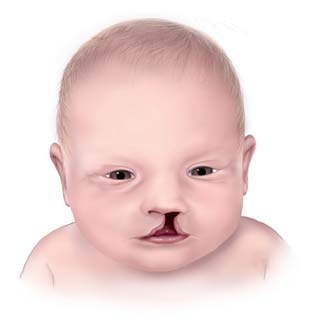
Illustrated baby with a cleft lip. Researchers at Yale University and UConn conducted a detailed genomic study that proved that epigenetic enhancers had a role to play in the development of cleft lips and/or cleft palates. (Source: Wikimedia Commons)
In addition, the team also reported that the enhancer-effects were most pronounced in the samples (which was made available to them) from embryos in the later stages of development.
Conclusions Drawn from the Research
This study indicates that cleft lip, palate, or both, may be caused by genomic defects, but in regions that are not involved in the expression of prevalent structural proteins. Rather, these mutations affect the complex and large-scale system of epigenetic enhancers that exist to ensure that embryonic development proceeds as normal.
It seems now that specific impairments that affect a relatively small fraction of the total craniofacial enhancer cohort may be significantly associated with the risk of this common facial disorder.
Therefore, this research may improve screening and treatment for this condition in the future.
Top Image: An incomplete, unilateral cleft lip. (Source: Felsir @ Wikimedia Commons)
References
A. Wilderman, et al. (2018) High-Resolution Epigenomic Atlas of Human Embryonic Craniofacial Development. Cell Reports. 23:(5). pp.1581-1597.
A. Kundaje, et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature. 518:(7539). pp.317-330.
WHO. (2003) Weekly epidemiological record: Global situation of the HIV/AIDS pandemic, end 2003. Wkly Epidemiol Rec. 50:(78). pp.425-432.
Blueprint for the Skull, 2018, UConnHealth News, https://today.uconn.edu/2018/05/blueprint-skull/, (accessed 4 May 2018)






No comment