There are thousands of known genes in the human genome. This number refers to the DNA sequences that code for actual proteins, which carry out functions in the body once expressed. These genes can, in turn, experience millions of variations, which may cause the said proteins to turn out differently than their original DNA "blueprint." In some cases, these abnormal proteins may be the basis of a disease state. Alternatively, genetic variations may cause a protein to become over- or under-expressed, with similar ramifications for health or longevity.
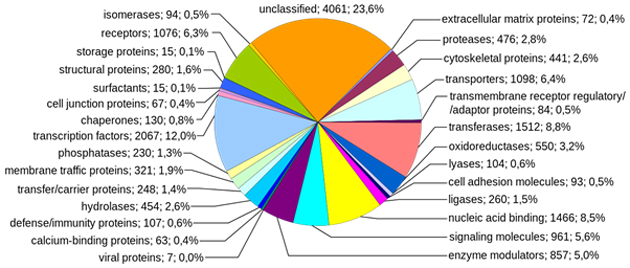
A recently-compiled breakdown of protein-coding human genes and what they do in the body. (Source: Häggström, Mikael @ Wikimedia Commons)
However, the human genome is not this simple as it also contains sequences that essentially do the ‘housekeeping’ for the rest of the genome.
This process may include the prevention of transcription, or the final stage of expression (in which the DNA template is converted to RNA and subsequently into the protein). The DNA, known as the regulatory code or regulatory space of the human genome, is also susceptible to mutations, which are associated with some illnesses.
The complex set of interactions that genes have with the rest of the coding genome makes it difficult to characterize, in terms of disease modeling, using conventional computing.
How the ML System ExPecto Works
However, in the age of AI, this situation is improving rapidly. An example in this new realm of variation-monitoring is ExPecto, a machine learning (ML) system developed at Princeton, in conjunction with the Simons Foundation in New York.
ExPecto has been trained to assess sequences with variations in the context of what the gene(s) in that sequence normally do. The program then extrapolates a variation’s effect on the protein (or regulatory action) that the gene in question codes. Accordingly, ExPecto predicts what that effect could eventually have on a phenotype (the biological and physiological manifestation of a gene).
ExPecto was designed and trained based on previous work that found convincingly causal relationships between disease markers in the genome and actual conditions. All the genome-wide association studies, available to the Princeton/Simons team, were included, on four specific conditions. These immunological health states were chosen to inform ExPecto’s algorithms and ability to predict disease-specific variants. Empirically-validated models of variations associated with the four conditions were also integrated into the system.
The AI was then applied to the problem of variations in regulatory genes (which selectively inhibit or promote the expression of different genes in response to different physiological circumstance) that are transcribed by human RNA polymerase II. This was done by simulating mutations in this area of the genome using the software.
ExPecto was able to identify over a 140 million of these variations, and the effect they would have on the phenotype during transcription. This result was attributed to the scalability of the AI.
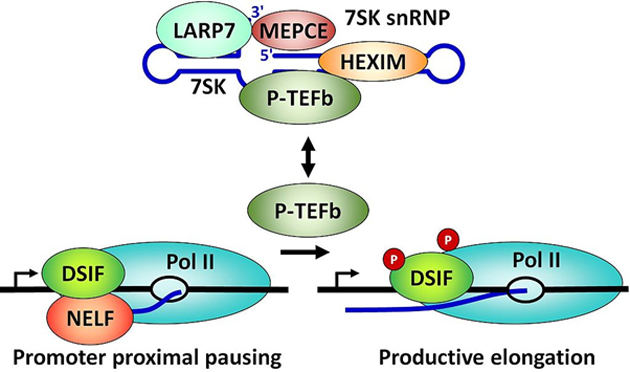
An example of transcriptional regulation. A mutation in one of these proteins (colored ellipses) could lead to abnormally high gene expression and, thus, a potential illness. (Source: David H. Price @ Wikipedia)
Great (ExPecto)tions: Applications in the Future
In other words, ExPecto was able to read a sequence, find the variations and extrapolate the effect of these on a (hypothetical) living human from scratch. Therefore, it may well be viable in the silico-system for disease and disease risk prediction.
The scientists from Princeton and Simons, led by Olga Troyanskaya (who holds positions at both institutes) also maintain that ExPecto could be used to model the effects of evolution, and of different evolutionary pathways, on the human genome. This is a reasonable assertion, as these processes are, at least, partially based on the acquisition of ‘favorable’ genetic variations, and, sometimes, on the loss of others.
All in all, this study suggests that ExPecto may be used to identify particularly dangerous mutations in the genome. This development could represent considerable advancements in health screening technology. Patients with these variations could be notified well-in-advance of the onset of an actual condition. ExPecto could also help assess different levels of relevant risks for different patients.
In addition, this ML system can keep track of potentially significant variations in the mutation-prone regions of the regulatory space of the human genome.
Top Image: DNA codes for the functional proteins that make up our cells, and more besides. (Credit: qimono @ pixabay.com)
References
J. Zhou, et al. (2018) Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nature genetics.
J. K. Pickrell, et al. (2010) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 464:(7289). pp.768-772.
A. Ramasamy, et al. (2014) Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 17:(10). pp.1418-1428.





No comment